Overview: A physics-guided deep-learning-based modular framework for segmenting tumors in oncological PET on a per-slice basis.
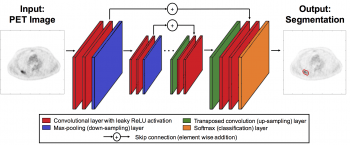
Software description: The software implements a three-module PET-segmentation framework to segmenting primary tumors in 3D FDG-PET images on a per-slice basis. Input to the software is a 2D image slice that contains a tumor. The software will output the 2D slice with the segmented tumor boundary.
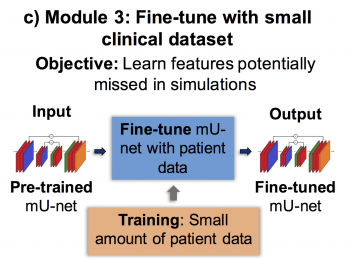
The first module generates PET images containing highly realistic tumors with known ground-truth using a new stochastic and physics-based approach, addressing lack of training data. The second module trains a modified U-net using these images, helping it learn the tumor-segmentation task. The third module fine-tunes this network using a small-sized clinical dataset with radiologist-defined delineations as surrogate ground-truth, helping the framework learn features potentially missed in simulated tumors.

In the first module, the software extracts tumor descriptors from clinical data to generate realistic simulated tumors. These will then be inserted to patient images through a projection-domain lesion-insertion approach. The workflow is as shown below.

In the second module, a modified U-net is trained using these simulated images, as shown through process below:

Finally, in the third module, the network is fine-tuned with a small-sized clinical dataset where manual segmentation is used as ground truth.
The software includes the complete architecture of the modified U-net, and implements the training-cross validation process as described above.
Software features: In our experiments, the developed software yielded reliable performance in both simulated (Dice scores: 0.87 (95% CI: 0.86, 0.88)) and patient images (Dice scores: 0.73 (95% CI: 0.71, 0.76)), outperformed several widely used semi-automated approaches, accurately segmented relatively small tumors (smallest segmented cross-section was 1.83 cm2), generalized across five PET scanners (Dice scores: 0.74), was relatively unaffected by PVEs, and required low training data (training with data from even 30 patients yielded Dice scores of 0.70).
Software and Data: Software and data are provided at this link.
Reference:
K. H. Leung, W. Marashdeh, R. Wray, S. Ashrafinia, M. G. Pomper, A. Rahmim and A. K. Jha, “A physics-guided modular deep-learning based automated framework for tumor segmentation in PET”, Phys. Med. Biol. (link)