Quantitative imaging for oncology is emerging as an important clinical and pre-clinical technique. The basic idea in quantitative imaging is to extract numerical features from medical images, and use these features for clinical decision making, for example to assess whether a patient with cancer is responding to therapy.
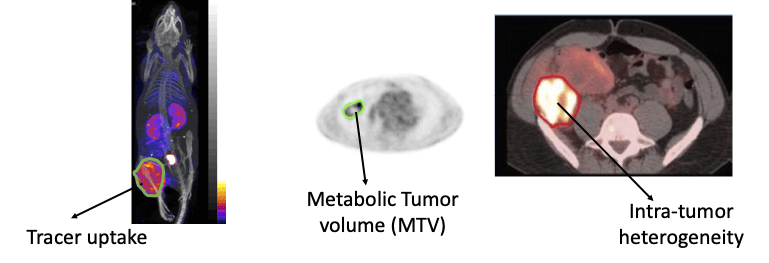
A key image-analysis step in measuring quantitative imaging features is image segmentation (see figure above). Currently, this segmentation is typically performed manually, which is erroneous and may lead to significant inter and intra-reader variability. Conventional semi-automated segmentation methods also suffer from limited reliability. Thus, there is a need to develop reliable segmentation methods. For this purpose, we have developed machine-learning-based approaches, including those based on deep learning. Recently, we developed a physics-guided deep-learning approach that yielded accurate performance in segmenting these images even when the number of training images was small. We also developed a new estimation-based segmentation methodology that, unlike conventional segmentation methods, estimates how much of a voxel in an image contains a certain region. We observed that this method outperformed a state-of-the-art AI-based PET segmentation approach. We are actively collaborate with oncologists and radiologists to translate these methods to the clinic, in the process demonstrating the value of our methods. 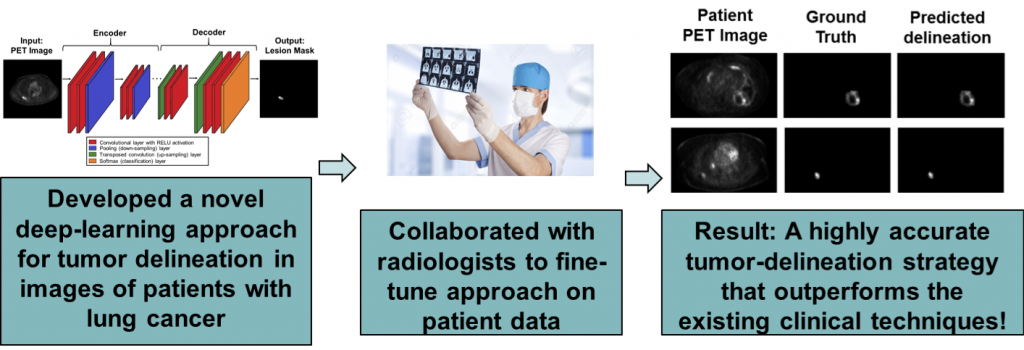
We have also developed techniques to evaluate these new quantitative PET methods with patient data without a ground truth. For more on this topic, see here.
References:
- Z. Liu, J. Mhlanga, R. Laforest, P. Derenoncourt, B. A. Siegel, and A. K. Jha, “A Bayesian approach to tissue-fraction estimation for oncological PET segmentation”, Phys. Med. Biol. 66 124002 (link) (WashU press release) (NIH press release)
-
F Yousefirizi, A K Jha, J Brosch-Lenz, B Saboury, A Rahmim, “Toward High-Throughput Artificial Intelligence-Based Segmentation in Oncological PET Imaging”, PET Clin. 2021 Oct;16(4):577-596 (link)
- Leung, W. Marashdeh, S. Ashrafinia, A. Rahmim, M. Pomper, and A. K. Jha, “A physics-guided modular deep-learning based automated framework for tumor segmentation in PET”, Phys. Med. Biol. (link) (arxiv) (press release)
- E. Mena, S. Sheikhbahaei, M. Taghipour, A. K. Jha, J. Xiao, E. Vicente, R. M. Subramaniam, “18F-FDG PET/CT Metabolic Tumor Volume and intra-tumoral heterogeneity in pancreatic adenocarcinomas: Impact of Dual-Time-Point and Segmentation Methods”, Clin. Nuc. Med., 2016.
- Mena, M. Taghipour, S. Sheikhbahaei, A. K. Jha, A. Yanamadala, R. M. Subramaniam, “Value of intra-tumoral metabolic heterogeneity and quantitative 18F-FDG PET/CT parameters to predict prognosis in patients with HPV-positive primary oropharyngeal squamous cell carcinoma”, Clin. Nucl. Med. 2016
- Leung, W. Marashdeh, S. Ashrafinia, M. Pomper. A. Rahmim, A. K. Jha, “A Modular Deep-Learning Based Fully Automated Framework for Tumor Segmentation in PET Images”, Phys. Med. Biol (link)
- A. K. Jha, E. Mena, B. Caffo, S. Ashrafinia, A. Rahmim, E. C. Frey and R. Subramaniam, “Practical no-gold-standard framework to evaluate quantitative imaging methods: Application to lesion segmentation in PET”, J. Med. Imag. (Special Section on PET imaging), 4(1), 2017 (pdf).
- E. Mena, M. Taghipour, S. Sheikhbahaei, A. K. Jha, L. Solnes, R. M. Subramaniam, “Value of intra-tumoral metabolic heterogeneity and quantitative 18F-FDG PET/CT parameters to predict prognosis, in patients with HPV-positive primary oropharyngeal squamous cell carcinoma”, Clin. Nuc. Med., 42(5); e227-34, 2017 (pdf)
- E. Mena, S. Sheikhbahaei, M. Taghipour, A. K. Jha, J. Xiao, E. Vicente, R. M. Subramaniam, “18F-FDG PET/CT Metabolic Tumor Volume and intra-tumoral heterogeneity: Impact of Dual-Time-Point and Segmentation Methods”, Clin. Nuc. Med., 42(1), e16-e21, 2017 (link)
- A. K. Jha, J. J. Rodriguez, A. T. Stopeck, “A maximum-likelihood method to estimate a single ADC value of lesions using diffusion MRI”, Mag. Res. Med., 76(6), 2016 (pdf)
- A. K. Jha, B. Caffo, E. Frey, “A no-gold-standard technique for objective assessment of quantitative nuclear-medicine imaging methods”, Phys. Med. Biol., 61(7), 2780-2800, 2016 (pdf)
- R. M. Stephen, A. K. Jha, D. J. Roe, et al., “Diffusion MRI with Semi-Automated Segmentation Can Serve as a Restricted Predictive Biomarker of the Therapeutic Response of Liver Metastasis”, Magn. Res. Imag., 33(10), 1267-33, 2015 (pdf)
- A. K. Jha, M. A. Kupinski, J. J. Rodriguez, R. Stephen, A. Stopeck, “Task-based evaluation of segmentation algorithms for diffusion-weighted MRI without using a gold standard”, Phys. Med. Biol., 57(13) 4425-46, 2012 (link)
- A. K. Jha, M. A. Kupinski, J. J. Rodriguez, R. M. Stephen, A. T. Stopeck, “Evaluating segmentation algorithms for diffusion-weighted MR images: a task-based approach”, Proc. SPIE 7627,76270L1-8, 2010 (Best Student Paper Award) (link)